CBBP Research Activities
The Computational Biology and Biological Physics Group, part of the Department of Astronomy and Theoretical Physics at Lund University, pursues a very broad spectrum of cross-disciplinary activities. In particular, the group develops and applies machine learning techniques to a variety of problems, develops models of macromolecules and cell systems, and investigates biological processes by statistical and and imaging techniques.
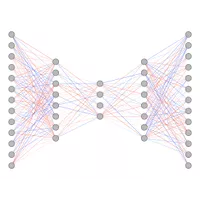
Machine learning
The machine learning research activities at CBBP cover both algorithmic development and a wide range of applications. The former is focused on neural network- and deep learning models, and includes development of methods for missing data imputation, survival analysis and generative models using variational autoencoders. Applications of machine learning focus mainly on biological and medical data, ranging from pure model building to understand biological processes or diseases, to very specific medical decision support tasks. All applications are carried out in close collaboration with the different partners. Examples of applications are analysis of proteomic and genomic data for early detection of cancer and autoimmune diseases, and detection of risk factors and optimal matching for heart transplantation. Some of the applications concern object identification and classification of images, ranging from satellite imagery to high resolution pathology images. |
Biomolecular physics
Biophysical properties of proteins, RNA and DNA have traditionally been studied mainly in dilute aqueous solution. Much less is known about biomolecular properties under cellular conditions. Basic phenomena that remain incompletely understood include the formation of membrane-less organelles (or biomolecular condensates), protein aggregation, and crowding-induced changes in properties such as protein stability.
Using methods from statistical and computational physics, we build models to investigate biomolecular processes and properties in dense systems. Some of the research is done in collaboration with experimental groups. On the computational side, we have a long-standing collaboration with researchers at the Jülich Supercomputing Centre.

Bionanophysics
The bionanophysics activities at CBBP involves several theoretical initiatives which combine bio- and nano-physics. One of the major directions of research is nanochannel-based optical DNA mappings. Such mappings serve as a fast way of providing both gene information, coarse-grained sequence and DNA length quantification of single plasmid and chromosomal DNA from bacteria and humans.
The group has close ongoing collaborations with experimental pioneers in the field and complements experiments by employing methods from statistical physics, computational physics, image analysis and statistics. Other bionanophysics projects pursued in the group include theoretical modeling of how confinement and crowding affect polymer and tracer particle dynamics.
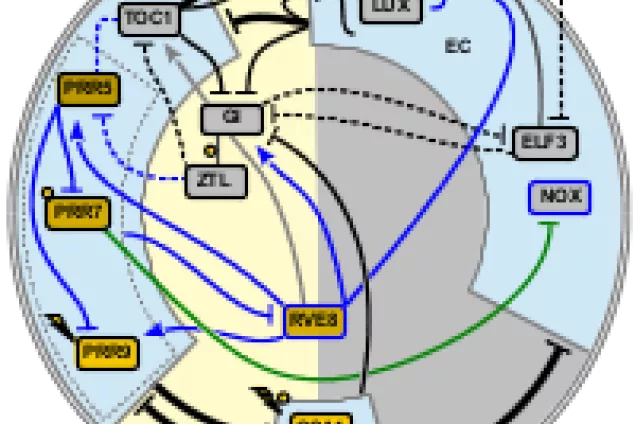
Systems biology
The systems biology research at CBBP is concerned with circuits of interacting genes and proteins that govern individual cell decisions. Computational models are developed to help understand and predict how genetic, epigenetic and metabolic circuits function dynamically in individual cells and multicellular systems. We conduct highly interdisciplinary research in close collaboration with experimental groups, developing modeling techniques linked to experimental methods and data.
Mammalian stem cell regulation is a major area for this research. Examples of topics include mechanisms controlling commitment of single T-cell progenitors, direct conversion of human adult fibroblast cells into neurons and microglial cells and reprogramming of skin cells into induced pluripotent stem cells. Non-mammalian topics include the regulation of extracellular oxidative degradation in ectomycorrhizal fungi.
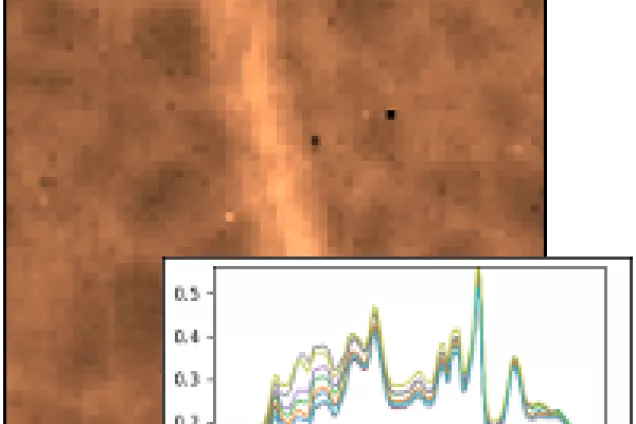
Spectroscopy methods
Microspectroscopic imaging generates vast amounts of data which need to be corrected and analyzed to reveal subtle chemical differences. The group is researching algorithms and developing software for processing images from vibrational spectroscopy, in close collaboration with spectroscopists and biologists. Tools are being developed for both IR and Raman data, and combining these techniques is of special interest.
Among the biological problems studied with these methods are the interactions between fungi and soil organic matter and protein aggregation in neurodegenerative disease. Specific examples include species-dependent alteration of cellulose crystallinity and spatial patterns of chemical modification of a lignin substrate by exploring hyphae.